Initial Benchmarks (Continued ..)
This is the continuation of previous post, and includes more benchmarks for different use cases. Last post concluded few initial benchmarks comparing KDtree, Cellgrid and Brute force method for two use case (1)Contact searches (2) Near neighbour selection. An obvious conclusion that cell list and KDtree are better alternatives as compared to brute force for large number of particles was quantified in terms of execution time. However, brute force is a better option for very less number of particles. Other data structures were also compared for single point queries and it was concluded that neighbour search as implemented in FATSLiM and Octree are viable candidates for distance calculations.
Since it was established that Octree and neighbour search in FATSLiM could be good replacements for fast distance evaluations for single point queries (fixed radius selection and near neighbour selections), a more detailed analysis of pair contact searches is covered in this post. For contact searches, grid based methods become more and more useful as usage of sparse matrix can become handy when the density is high as opposed to traversing the tree to identify each particle’s node in tree based data structures. This usage of sparse matrix reduces the query time, which becomes a huge time saving factor for large number of particles. As a next step to previous benchmarks, the Cellgrid package is optimized and used here to search for the contact pairs(see here). As a modification, a optional argument was added to the capped_self_distance_array which calculates the cellsize corresponding to 30 particles per cell assuming a uniform distribution of particles. The cellsize has a lower bound of cutoff distance, since decreasing the cellsize below the cutoff distance would require to extend the search to cells beyond the immediate neihbourhood of the query point.
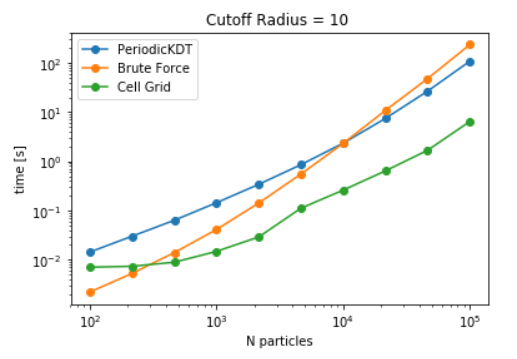
CellGrid package, KDTree and brute force methods are first compared for the use case to find all the pairs within a fixed distance (see here). For brute force calculations, distance_array is used in place of self_distance_array due to its high memory consumption. To avoid the repetatitve calculations due to each pair, care is taken to evaluate distance only once for each pair. Similarly, individual queries is pruned in KDtree and a list with all the pairs is maintained. For the case of Cellgrid, a distance matrix containing the particles within its neighbourhood is constructed. This matrix is masked based on the distances. To study the scaling behaviour of all three methods, a cutoff distance of 10 distance units is fixed and execution time is registered for different particle densities in a fixed box.
Time to evaluate particle within a distance of 10 distance units
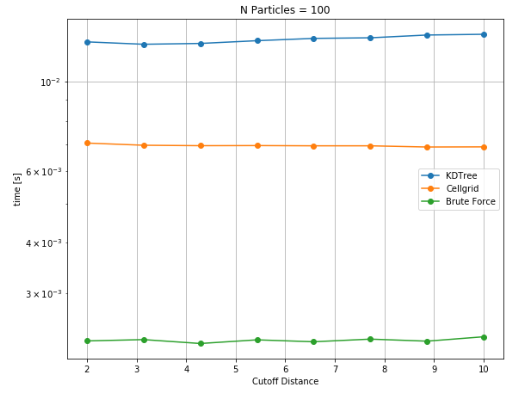
Since, cutoff radius is an important factor for performance, similar studies were performed to characterize the effect of cutoff radius for different particle densities. Although the trend with cutoff distance is continuously increasing for complete range of particle density i.e. for number of particles ranging from 100 to 100k. However, a significant relative shift can be seen from low to high number of particles. While brute force takes notably less time as compared to other data structures, the transition to cellgrid becomes evident at higher particle density.
Variation of execution time for different cutoff distances for 100 particles
Variation of execution time for different cutoff distances for 17k particles
Variation of execution time for different cutoff distances for 100k particles
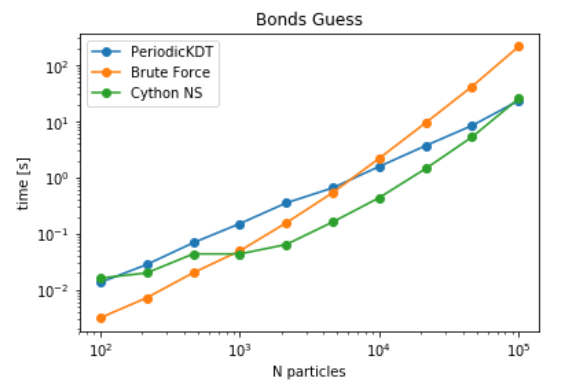
It can be seen that Cell-lists become advantageous as the number of particles increase. Since FATSLiM have a more efficient implementation of grid search, The next step is to check the timings of PBC aware Neighbour search module for pair contact searches. For this benchmark, we chose a particular implementation of bonds_guess, which is implemented in MDAnalysis at MDAnalysis.topology.guessers. The goal of this function is to identify the bonds between atoms by identifying the neighbouring atoms and checking the distance between them relative to the sum of their radius. Current implementation evaluates all the pair contacts for each particle using naive distance algorithm. It is anticipated that this algorithm is very costly and can be replaced with other data structures to improve the performance. It can be seen that while tree and grid structures both are advantageous at very large particle densities, Neighbour search (linear grid search) is more advantageous at intermediate particle densities as well. As expected a transition from brute force to KDTree/ Cell-List is achieved at lower particle densities i.e. around 1k for cell-list and around 6k for Periodic KDTree (see here).
Benchmarks for Guessing bonds between atoms in a static dataset of atoms
Apart from the increase in performance, another trick which only calculates distances from half the neighbours (as implemented in Cellgrid) is not implemented in Neighbour search routine. It is anticipated that performance of this method can be further increased by adopting this procedure.
In conclusion, for fixed radius neighbor selection, cython optimized neighbour search routine is a robust algorithm which can be used in tandem with brute force method and /KDTree in few cases depending on the application. Another recommendation would be to switch from Biopython KDtree to scipy cKDTree, which is found to be faster than the former(see here). The order of magnitude depends on the query point, but a performance improvement of the order of 5 times is observed as a best case i.e. for uniform distribution. For contact searches, it is already shown in this blog that, a cutoff of around 1000 particles can be used to switch between brute force and neighbour search routine from FATSLiM. Modified script for the neihbour search routing can be found here.
Thanks to Sébastian to introduce the cython optimized Neighbour search routine along with the benchmarks (see here).
Till Next Time!!!